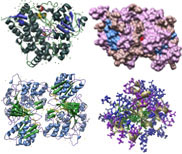
Cytochrome P450 Images
-
Consensus Matching of 4 Cytochrome P450s as ribbon-structures. P450s and Colors: CYP3A4 [1TQN, Homo sapiens], purple; CYP2B4 [1PO5, Oryctolagus cuniculus], blue; CYP2C5 [1DT6, Oryctolagus cuniculus], rose; CYP2C9 [1R9O, Homo sapiens], green.
-
Consensus Matching of 4 Cytochrome P450s as ribbon-structures with bound ligands. P450s: CYP3A4 [1TQN, Homo sapiens], CYP2B4 [1PO5, Oryctolagus cuniculus], CYP2C5 [1DT6, Oryctolagus cuniculus], and CYP2C9 [1R9O, Homo sapiens]. Color key: P450 backbone structures, blue; K-helix consensus, brown; PERF consensus, yellow; heme-binding domain, orange; bound ligands ( heme [all], flubiprofen [CYP2C9], glycerol [CYP2C9], Samarium ion [CYP2C5], Sulfate ion [CYP2C5]), pink.
-
Backbone line structure, ribbon consensus sequences, and bound ligands, in 3 views: View #1, View #2, View #3. P450 color key: CYP3A4 [1TQN, Homo sapiens], purple; CYP2B4 [1PO5, Oryctolagus cuniculus], blue; CYP2C5 [1DT6, Oryctolagus cuniculus], rose; CYP2C9 [1R9O, Homo sapiens], green. Feature color key: K-helix consensus, brown; PERF consensus, yellow; heme-binding domain, orange; bound ligands bound ligands (heme [all], flubiprofen [CYP2C9], glycerol [CYP2C9], Samarium ion [CYP2C5], Sulfate ion [CYP2C5]), pink.
-
CYP3A4 [1TQN, Homo sapiens] ribbon detail. Color key: helix structures, gray; strand structures, blue; PERF consensus, green; K-helix concensus, purple; heme-binding consensus, orange; heme ligand, yellow.
-
Consensus matching of CYP83B1 [Arabidopsis thaliana] as predicted by CPHmodels 2.0 to CYP2C5 [1DT6, Oryctolagus cuniculus], displayed in ribbon and ball-and-stick detail with bound ligands. Feature color key: CYP83B1, green; CYP83B1 PXRX, K-helix, and heme-binding regions, red; CYP2C5, blue; CYP2C5 PXRX, K-helix, and heme-binding regions, pink; bound ligands (heme, Samarium ion, Sulfate ion [all bound to CYP2C5]), yellow.
Glycosyltransferase Images
-
UDP-Glucosyltransferase Gtfb [1IIR] from Amycolatopsis orientalis, ribbon-structure with ligand. Color key: helix structures, green; strand structures, purple; bound ligands (Sulfate ion, Magnesium ion), yellow.
-
UDP-Glucosyltransferase Gtfb [1IIR] from Amycolatopsis orientalis, ball-and-stick structure with ligand. Color key: helix structures, green; strand structures, purple; bound ligands (Sulfate ion, Magnesium ion), red.
-
UDP-Glucosyltransferase Gtfb [1IIR] from Amycolatopsis orientalis, ribbon and ball-and-stick structure with PROSITE consensus sequence. Color key: main structure, grey; PROSITE consensus, green ball-and-stick.
-
UDP-Glucosyltransferase UGT85A4 from Arabidopsis thaliana as predicted by 3D Jigsaw and AL2TS, consensus matching. UGT71G1 [2ACV, Medicago truncatula] was used as the template for both predictions. Note the similarities and differences in the predicted structures. Color key: 3D Jigsaw prediction, blue; AL2TS prediction, purple.
-
Consensus matching of UDP-Glucosyltransferase UGT85A4 from Arabidopsis thaliana predicted structure by AL2TS and UGT71G1 [2ACV] from Medicago truncatula. Ribbon detail with UPD-glucose bound to UGT71G1 and plant-UGT motif. Color key: UGT85A4, main structure, blue; UGT71G1, main structure, purple;UDP-glucose, yellow; PROSITE consensus, both UGTs, red.
-
Consensus matching of UDP-Glucosyltransferases UGT85A4 from Arabidopsis thaliana (predicted structure by AL2TS), UGT72B5 from Lotus japonicus (predicted structure by AL2TS), and UGT71G1 [2ACV] from Medicago truncatula against a UDP-glucose:flavonoid 3-O-glucosyltransferase [2C1X] from Vitis vinifera as edged ribbon structures. Color key: UGT from grapewine, yellow; UGT85A4, green; UGT71G1, blue; UGT72B5, purple.
-
Grapewine UDP-glucose:flavonoid 3-O-glucosyltransferase [2C1X, Vitis vinifera], wireframe detailf. Color key: helix, blue; strand, pink; backbone, beige; PROSITE consensus, dark purple.
β-glucosidase Images
-
Cyanogenic Betaglucosidase [1CBG] from Trifolium repens, ribbon-structure with water. Color key: helix structures, purple; strand structures, red; backbone, green; water, light purple.
-
Consensus matching of Cyanogenic Betaglucosidase [1CBG] from Trifolium repens and Dhurrinase 1 [1V02] from Sorghum bicolor, ribbon structures. Color key: Dhurrinase 1 [1V02], blue; White clover Cynogenic BGD, purple.
-
Putative Betaglucosidase from Oryza sativa, structure predicted by AL2TS, solid surface structure. Color key: helix, grey; strand, purple; backbone, green.
Cytochrome P450 Reductase Images
-
NADPH Cytochrome P450 Reductase [1AMO, Rattus norvegicus], ribbon structure with ball-and-stick ligand. Color key: helix, blue; strand, green; bound ligands (FAD, FMN, NAP), yellow.
-
NADPH Cytochrome P450 Reductase [1AMO, Rattus norvegicus], mesh surface structure. Color key: helix, green; strand, pink; backbone, dark grey.
Cytochrome b5 and b5 Reductase Images
-
Cytochrome b5 [1B5B, Rattus norvegicus] ribbon structure. Color key: helix, blue; strand, gold.
-
Cytochrome b5 [1B5M, Rattus norvegicus] mesh surface structure with ball-and-stick ligand. Color key: helix, green; strand, gold; bound ligand (heme), red.
-
Cytochrome b5 reductase [1NDH, Sus scrofa]
ribbon structure. Color key: helpix, purple; strand, green; bound ligand (FAD), blue.
|